DoCM, the Database of Curated Mutations, is a highly curated database of known,
disease-causing mutations that provides easily explorable variant lists with direct links to source
citations for easy verification.
Publications are identified for curation by disease experts and through review of published lists and resources that outline important cancer mutations. Mutations outlined in the literature are included in DoCM if they satisfy the criteria listed below.
Clinical evidence
Functional evidence
Using context clues of the citing literature, useful metadata is obtained from public datasets to uniquely identify a mutation (such as genomic position and transcript), map the mutation to a disease (using the disease ontology), and catalogue other useful information.
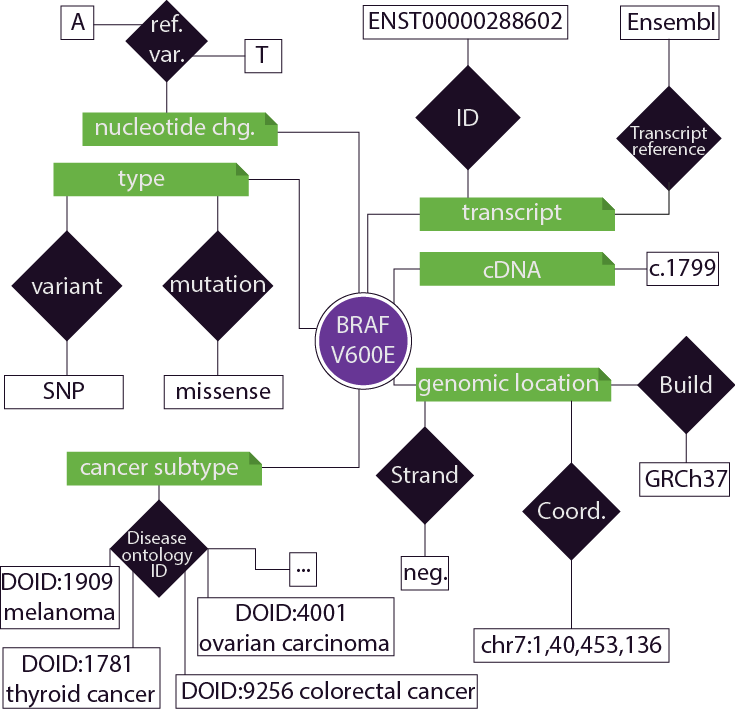
The following is an anecdotal example of the curation involved for the variant BRAF V600E. Typically the literature only lists the gene and amino acid change, requiring extensive curation to uniquely identify the variant. Correct genomic coordinates on a consistent genome build need to be identified, with accompanying nucleotide information. Occassionaly there are multiple nucleotide changes that are synonymous with a particular amino acid change. A representative transcript that correctly models the mutation described in the literature also needs to be specified. Cancer subtypes are specified using the disease ontology nomenclature. Tags can be added to an individual variant to provide useful metadata, examples include "pathogenic", "functional mouse model", "prognostic", and/or "activating".
Variants should be grouped into batches by commonalities like disease or mutation type if curated directly from the literature. Batches can also be created based on a publically available listing of variants that is in scope, like My Cancer Genome or the Drug Gene Knowledge Database. Batches can be submitted, following the batch submission instructions, on the batch submission page. Curators should annotate their curation process and explain the reasoning for including a batch into DoCM in the batch rationale statement on the submission form. This statement allows more transparency to the curation process and allows DoCM users to better understand why variants were included.
Following submission of a batch, the DoCM web app automatically annotates the variant using VEP, validates the the publications pubmed ids using PubMed, and validates the disease ontology ids using the disease ontology API. After annotation and validation, the variants are reviewed by the moderators listed on DoCM’s about page. DoCM moderators ensure that the a submitted batch contains no errors in annotation and validation, that the batch is in scope of the resource, and that the variants appear to be referenced in the listed literature. The moderator will start a dialogue with the submitter via email to correct any errors/discrepancies and then accept or reject the variants in the batch. These variants are then staged for inclusion in DoCM and upon submission of multiple batches the moderator can create a new version of the database.
DoCM by The McDonnell Genome Institute at Washington University School of Medicine is licensed under a Creative Commons Attribution 4.0 International License. Questions? Comments? Concerns? You can contact us here.